Research in the lab is generally related to the molecular evolution and cell biology of microbial eukaryotes, protists (i.e. eukaryotes that are not animals, fungi, or plants). Protists are mostly single celled organisms, but are many are extremely complex and sophisticated despite their small size. Protists also represent the greatest part of eukaryotic diversity, although most protists groups are very poorly studied, especially at the molecular level. We use molecular biology, microscopy, and genome wide analyses to study a number of questions in different lineages of protists. Since most protists are not cultivated, we also use new methods to get important species into culture and single-cell genomics and transcriptomics methods to work on those we cannot culture. Major themes of our work are symbiosis and parasitism, as well as the diversity and roles of heterotrophs.
One major theme in the lab is symbiosis and especially the origin and evolution of cellular organelles, mitochondria and plastids. Symbiosis is when two species live in close association, and we are in particular interested in endosymbiosis, or when one cell lives inside another. This is often taken as being of mutual benefit, but it is becoming less clear that endosymbiotic systems ever really live together mutualistically. We are looking at a variety of systems, both very ancient and very new, to try to understand how these relationships form and what affect they have on the cells involved. The organelles are a special case. They are highly integrated parts of the eukaryotic cell that originated by the uptake and retention of a bacterium. In the case of mitochondria this involved an alpha-proteobacterium and took place around the time of the origin of eukaryotes. Plastids originated more recently from a cyanobacterium. Plastids have also spread between eukaryotic lineages by a process called secondary endosymbiosis. In this case, a plastid-bearing alga is itself taken up by another eukaryote, and its photosynthetic apparatus is retained by this new, secondary, host. These are special cases because they have traceried genes to their host, the products of which are now targeted back as proteins, and their impact on eukaryotic evolution is profound.
Another major focus of the research is the transition of free-living organisms to parasitism and how this affects their organelles and metabolism, in particular in the case of intracellular parasites. This is in many ways related to the study of endosymbiosis, since the relationship between the host and guest share many features in common, but the context is clearly different. It also crosses over because our main subject are the apicomplexan parasites, and one of the key features we look at are their plastids. Apicomplexans are a group of intracellular parasites that include several major disease-causing agents, such as Toxoplasma, Cryptosporidium, and the malaria parasite, Plasmodium. In the mid-1990s, apicomplexa were surprisingly shown to contain a plastid, although they are obviously not photosynthetic. The plastid function is now fairly well-characterised in model parasites, where genes for nuclear-encoded plastid targeted proteins involved in the biosynthesis of heme, fatty acids, and isoprenoids have been found. We have been working on the origin of this plastid and its metabolic enzymes for many years, and also the distribution of the plastid in extant apicomplexa and their close relatives. Many of these relatives have been found in some association with coral, and this has in turn led us to work on the diversity of coral-associated protists and how they affect our understanding of the early branches of the apicomplexan tree and the evolution of characters involved in parasitism.

Doughnut biodiversity, and the origin of doughnuts are questions of great biomedical importance that have vexed scientists for many years. We have addressed these problems using a novel environmental sampling technique that simultaneously examines doughnodiversity and the relative abundance of different species in the doughnosphere. Based on the hypothesis that doughnut expression will leave environmental traces, doughnodiversity was sampled from the sub-rimal regions of used double-doubles. Double-doubles were procured at Spring Garden Road, Halifax, and rims were rolled-up manually, exposing traces of doughnofauna in the rim core. In total, 10,000 rim cores have been examined between the months of April and May (we are indebted to the members of the Doolittle Lab for field work). DNA was prepared from isolated rim-doughnofauna, and used as templates in environmental PCR reactions for the Surface Coat Sugar gene, SGS. Sequences were found to fall into three major families, which were identified by immunogold labeling and electron microscopy as representing the classical ring-form doughnut, or Ringozoa, the jelly- and cream-filled forms, or Jellyzoa, and the more recently discovered and putatively primitive Timbitozoa. Within each family the relative abundance of species varied dramatically, with Honi krulleri being the most abundant Ringozoa, and Bostonia creami being the most abundant Jellyzoa. Interestingly, an unexpected abundance of Timbitozoa was discovered in the relatively benign environment of Spring Garden Road, despite the widespread belief that Timbitozoa are restricted to extreme environments such as toddlers' birthday parties, school staff meetings, and of course, campus security stations. Unexpectedly, we also found that the Surface Coat Sugar gene is the product of an ancient gene duplication, resulting in "glazed" and "dusted" phenotypes. Since Ringozoa, Jellyzoa, and Timbitozoa all contain both sugar-glazed and sugar-dusted varieties, we were able to root the universal doughnut tree using this ancient sugar paralogy. Astonishingly, we found Timbitozoa to be sisters to the Jellyzoa, indicating that the Ringozoa are the most primitive family of doughnuts. Clearly, the Timbitozoa are not primitively simple doughnuts, but instead are secondarily derived, perhaps the result of extreme marketing pressure. This finding focuses new importance on the study of the Ringozoa, and also on the characters shared among the three families. We suggest a number of shared characters exist between Jellyzoa and Ringozoa would have been present in the last common ancestor of doughnuts, which we call the "Proughgenut".

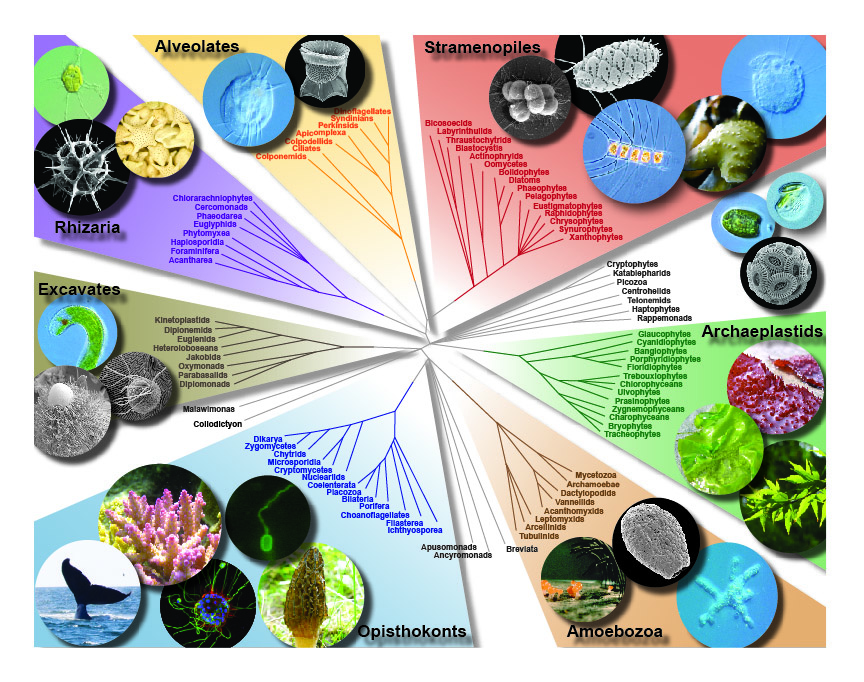
Most of our work relies on an accurate and well-sampled phylogeny of eukaryotes, and this is in itself a major goal of the lab since it guides so many evolutionary and ecological inferences. The phylogeny of eukaryotes is a huge puzzle and we are contributing to its piecing together using ‘phylogenomic’ analyses, or analysing the tree using hundreds of genes gathered from genomic and transcriptomic data. These trees, from our lab and from others, are converging on a view of eukaryotic phylogeny being made up of a small number of very large and diverse eukaryotic “supergroups”. Supergroups are something beyond the traditional kingdoms, and are all dominated by microbial eukaryotes (i.e. animals, fungi, and plants all fall within one of these supergroups, and all have different microbial relatives). Another part of this problem is adding new taxa to the tree of eukaryotes. Eukaryotic diversity is huge, and we simply don’t know about most of it. In particular, relatively major lineages are still being discovered surprisingly often. We are focusing on these missing taxa, and in particular ones that branch in “empty” parts of the tree, or near branches where evolutionary major transitions (like those described above) were taking place. These new lineages often change the tree or support for important parts of the tree, and also hold keys to reconstructing major evolutionary events. But if they are hard to culture or rare in nature, they can be overlooked even with huge molecular surveys of microbial diversity. We use classical methods like culturing and microscopy, together with single cell genomic and transcriptomics to try to fill in some of these gaps.