
CAN-TILL Projects

 |
CAN-TILL Projects
|
 |
 The major CAN-TILL project
at this time is focussed on the oilseed crop Brassica napus For canola TILLING
data please go here:
The major CAN-TILL project
at this time is focussed on the oilseed crop Brassica napus For canola TILLING
data please go here: Past
projects include Ecotilling in Populus
trichocarpa (black cottonwood), and TILLING in Arabidopsis, the vegetable crop
Brassica oleracea, and in the
soil nematode Caenorhabditis elegans.
|
|
| Brassica napus | |
| Arabidopsis | |
| Brassica oleracea | |
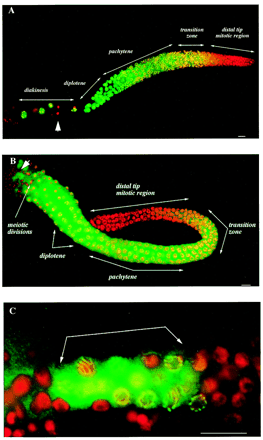
| Caenorhabditis elegans | |
| Populus trichocarpa (Ecotilling) |
 The Brassica
project was initially funded as part of two oilseed
initiatives funded by Genome Canada/Genome Alberta (Designing
Oilseeds for Tomorrow's Markets), and AVAC (Bioactive Oils Programme), and subsequently through funding
from the Alberta Innovates Phytola Centre led by Dr. Randall
Weselake. The goals of these projects were to use TILLING to identify endogenous mutations that
affect oil content, seed coat characteristics and levels of
anti-nutritional factors. It is anticipated that the results of
this research will enhance the overall usefulness of canola
seed, leading to improved meal for food and animal feed
applications, and diversified seed oil content for nutritional
and industrial uses. We have generated a population of
approximately 3000 EMS-mutagenised B. napus lines for
TILLING, and have identified mutations in a number of genes
requested by researchers. The mutation rate for our population
is approximately 1 mutation per 100Kb. This should allow the
identification of several null or deleterious alleles for each
gene screened.
The Brassica
project was initially funded as part of two oilseed
initiatives funded by Genome Canada/Genome Alberta (Designing
Oilseeds for Tomorrow's Markets), and AVAC (Bioactive Oils Programme), and subsequently through funding
from the Alberta Innovates Phytola Centre led by Dr. Randall
Weselake. The goals of these projects were to use TILLING to identify endogenous mutations that
affect oil content, seed coat characteristics and levels of
anti-nutritional factors. It is anticipated that the results of
this research will enhance the overall usefulness of canola
seed, leading to improved meal for food and animal feed
applications, and diversified seed oil content for nutritional
and industrial uses. We have generated a population of
approximately 3000 EMS-mutagenised B. napus lines for
TILLING, and have identified mutations in a number of genes
requested by researchers. The mutation rate for our population
is approximately 1 mutation per 100Kb. This should allow the
identification of several null or deleterious alleles for each
gene screened.Photo
courtesy of Canola
Council of Canada
For canola TILLING data please go here: B.
napus TILLIG data (or email
erin.gilchrist@ubc.ca).
Link to publications arising from this project:
A past project, TILLING for mutations in
the vegetable crop species Brassica oleracea,
identified mutations that are thought to play a role in
plant response to abiotic stresses that particularly
affect crop yield in Canada. These include elements such
as temperature, drought and nutrient imbalance. Phase one
of this project involved TILLING in Arabidopsis
homologues of Brassica genes that were thought to
be important in abiotic stress responses in these crop
species. The second phase of the project involved TILLING
in Brassica oleracea, a species closely related to
the commercially important crop, Brassica napus.
Photo from Brassica genetics for the classroom

 The first tree to have it's
genome sequenced is the western black cottonwood, Populus trichocarpa. This tree has a natural
range that spans from Alaska to southern California, and
from the Pacific Coast into interior mountain ranges in
British Columbia, Washington and Oregon. As a first step
towards analyzing genetic variation in this species, a live
reference collection of P. trichocarpa has been established at the University of British
Columbia (UBC) that includes trees from more than 140
different populations (see map).
The first tree to have it's
genome sequenced is the western black cottonwood, Populus trichocarpa. This tree has a natural
range that spans from Alaska to southern California, and
from the Pacific Coast into interior mountain ranges in
British Columbia, Washington and Oregon. As a first step
towards analyzing genetic variation in this species, a live
reference collection of P. trichocarpa has been established at the University of British
Columbia (UBC) that includes trees from more than 140
different populations (see map). Link to publications
arising from this project: 
This project was a collaboration with Quentin Cronk (Centre for Plant Research and Botanical Garden, UBC) and the Genome BC Forestry Genomics project.


 Through collaboration with the Seattle TILLING Project, we used the model plant Arabidopsis thaliana as a tool for examining plant
gene function through reverse genetics. We were able to
provide researchers in the Genome Prairie Abiotic Stress
group and the Genome B.C. Forestry Genomics group with a
total of 171 mutations in 20 different genes that may be
commercially important in agriculture or forestry. This
project is completed.
Through collaboration with the Seattle TILLING Project, we used the model plant Arabidopsis thaliana as a tool for examining plant
gene function through reverse genetics. We were able to
provide researchers in the Genome Prairie Abiotic Stress
group and the Genome B.C. Forestry Genomics group with a
total of 171 mutations in 20 different genes that may be
commercially important in agriculture or forestry. This
project is completed.
