Species Discovery & Evolutionary History
The
long-term objectives of our research program are to:
- Discover and characterize novel traits associated with locomotion, feeding and symbiotic interactions;
- Reconstruct the evolutionary histories of complex ultrastructural systems (e.g., ocelloids and nematocysts) using comparative molecular methods and high-resolution microscopy;
- Accelerate the discovery and characterization of new and poorly known lineages of (uncultured) organisms from diverse marine environments;
- Understand major trends and reoccurring innovations in eukaryotic evolution (e.g., convergent evolution over vast phylogenetic distances).
More focused research projects are motivated by these research themes.
Commentaries about the lab’s work on marine meiofaunal animals:
- Research overview starring postdocs Maria and Niels in Hakai Magazine (2017): Tiny dragons between grains of sand.
(Video)
- The micro monsters beneath your beach blanket.
(Link)
- What does it take to live between the grains of sand?
(Link)
- How to extract meiofauna from marine sand: The bubble and blot method
(Video)
- Research overview in UBC's Frontier Journal: Evolution Revolution
(PDF)
Research on the diversity of eukaryotes is exploratory and progresses in four principal phases:
- Collection of the organisms from diverse habitats (e.g., San Diego - California, Hakai Institute - Calvert Island, Bamfield Marine Sciences Center - Vancouver Island, and Curacao - Caribbean);
- Identification, specimen preparation and characterization of the organisms using high resolution microscopy;
- Isolation of the organisms for transcriptomics and genomics;
- Computational analyses associated with digital image processing and comparative molecular biology.
Outcomes of this research accelerate the discovery and characterization of new and poorly known species of eukaryotes from diverse environments and identify novel ways in which these organisms have solved basic biological problems, such as locomotion, nutrition, and photoreception.
Evolution of Complex Organelles
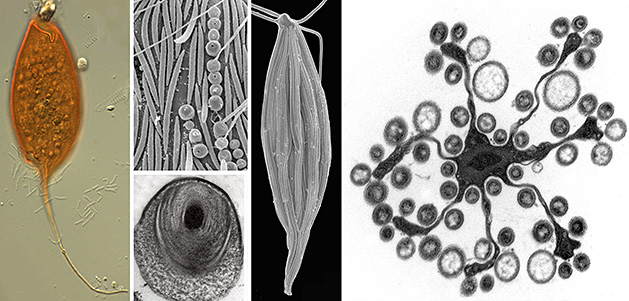
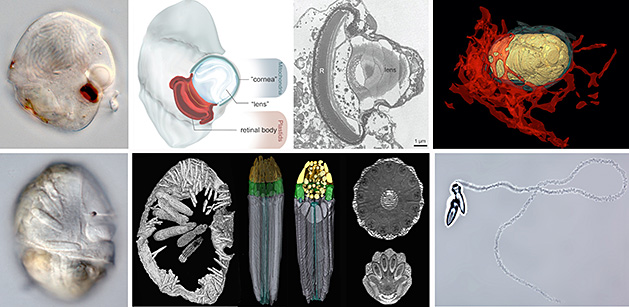
Comparative studies of single-celled eukaryotes, such as dinoflagellates, offer idiosyncratic systems for tracing the intermediate stages that gave rise to highly integrated subcellular systems. For instance, some dinoflagellates have organelles called ocelloids that look like the subcellular analogues to camera eyes in cephalopods and vertebrates (shown in the images below); some dinoflagellates have complex harpoon-like organelles that look very similar to the nematocysts of cnidarians or the colloblasts of ctenophores.
Our research addresses the ultrastructural and genomic diversity of many different groups of marine dinoflagellates with complex morphologies, such as dinophysoids, warnowiids and polykrikoids, in order to understand how different groups of eukaryotes bend the rules of biology far beyond what we see in the better-studied (model) eukaryotes.
Marine Parasitology
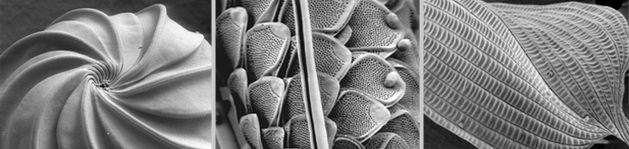
Marine gregarines (a few examples of their feeding stage are shown in the SEM images below) are a diverse and poorly understood group of single-celled parasites of invertebrate hosts and provide important insights into one of the most enigmatic evolutionary transformations in the history of life: the origin of obligate intracellular parasites from free-living, photosynthetic ancestors. Apicomplexans are a group of single-celled parasites that are closely related to dinoflagellates, and despite being infamous as pathogens of humans and livestock (e.g.,
Plasmodium, the causative agent of malaria), most of apicomplexan diversity is thriving in the world's oceans. The evolutionary origin of apicomplexan parasites began within marine sediments; free-living relatives to apicomplexans are tiny (< 10 microns) vampire-like predators, called “colpodellids”, that feed on other microbial eukaryotes. Some marine gregarines share this same mode of feeding and are inferred to have retained several ancestral traits for apicomplexans as a whole (e.g., extracellular feeding stages, a lifecycle consisting of a single host, and a prevailing presence in marine environments).
Because of their widespread presence in animals and a long history of being overlooked by the scientific community, it is expected that the vast majority of gregarines lineages (likely in the millions) remains unknown. Very few gregarines have been examined at the ultrastructural level and only a handful of gregarines have been studied at the molecular level. Addressing this black hole of basic (correlative) knowledge is among the most ambitious components of our research.
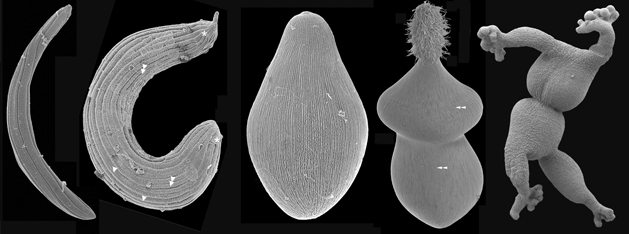
Biodiversity Between Grains of Sand
Our research explores the diversity and evolutionary history of traits in marine meiofaunal organisms, including several very distantly related lineages of eukaryotes (e.g., euglenozoans, dinoflagellates and cryomonad cercozoans) and miniaturized animals (e.g., scalidophorans and turbellarians). The lab is interested in the broad patterns and processes of convergent evolution associated with this lifestyle and how lineages of mieofuanal animals, single-celled eukaryotes and bacteria living in the same environments interact with each other. Some current research projects in the lab are focused on the biology and overall diversity of turbellarians and scalidophorans (i.e., kinorhynchs, loriciferans and priapulids) using confocal microscopy, FIB-SEM, transcriptomics and DNA barcoding. Many meiofaunal animals have co-evolved with symbionts and have acquired extremely reduced anatomical/physiological traits associated with miniaturization.
Other projects are focused on characterizing the traits of euglenids (left-most image shown below), dinoflagellates (second and third images from the left, shown below) and cryomonad cercozoans (two right-hand images shown below), which are diverse and poorly understood groups of microbial eukaryotes that inhabit marine sediments. Although all three groups have adopted similar modes of predatory life and thrive as part of the same microbial communities within marine sand, they could not be more distantly related to one another: euglenids are part of a supergroup called the Excavata; dinoflagellates are part of a supergroup called the Alveolata; and cryomonads are part of a supergroup called the Rhizaria. Despite these vast phylogenetic distances, some members within each of these groups look remarkably similar to one another at many different levels. For instance, all three of these groups have independently acquired a large and diverse suite of ultrastructural features, such as sophisticated feeding apparatuses, modes of gliding locomotion, extrusive organelles, robust cell surfaces, permanently condensed chromosomes, and in some, plastids via secondary endosymbiosis.
Improved knowledge of the ultrastructural diversity and phylogeny of these groups will provide evidence necessary for understanding cellular adaptation within interstitial habitats, complexity, and endosymbiosis. This research will also bring into the forefront the cellular context needed for interpreting the huge amount of genomic data being generating around the world, especially environmental PCR sequencing surveys of microbial diversity.
The lab continues to investigate the diversity of meiofaunal animals, euglenids, dinoflagellates and cryomonads in order to:
- Discover and characterize innovations associated with locomotion, photoreception and feeding;
- Provide well characterized reference taxa needed for accurate interpretations of environmental DNA surveys;
- Understand the adaptive relationship between cytoskeletal diversity and the secondary endosymbiotic origin of photosynthesis.
Because the vast majority of predatory flagellates are uncultivated and have only been minimally described with line drawings using light microscopy, there is enormous potential for ultrastructural discoveries using single-cell electron microscopy (e.g., SEM, TEM and FIB-SEM) that have implications for cell biology, developmental biology, evolutionary biology and paleontology. All of these data will be placed in a molecular phylogenetic context in order to infer trends in the evolution of morphological traits and the arms races that exist between single-celled predators and their prey.
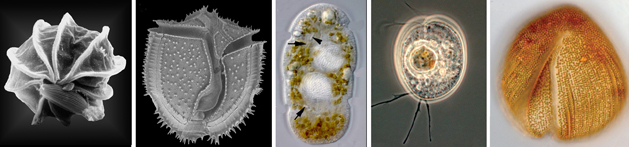
Symbiotic Interactions
Microbial eukaryotes in low oxygen environments are often enveloped with bacteria, but the diversity and functional significance of these relationships are poorly understood. The lab is continuing to characterize a distinct subgroup of euglenozoans, namely the Symbiontida (e.g.,
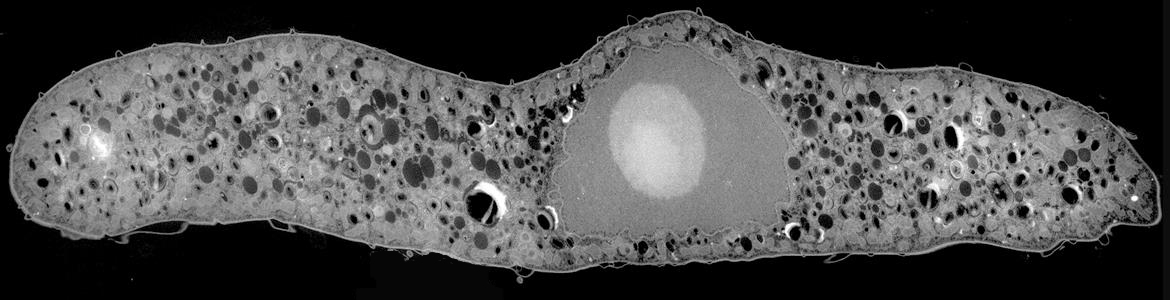
Calkinsia -- orange cell on the left side of the images below -- and
Bihospites), from oxygen-depleted and sulfidic marine environments. By definition, all members of this group carry epibionts that are intimately associated with underlying modified mitochondrion beneath the surface of the hosts (i.e., putative hydrogenosomes). We have shown that some of these lineages have additional ultrastructural traits that facilitate these epibiotic interactions, such as a complex and stable extracellular matrix consisting of conduits between the host cell and the enveloping epibionts.
Our ultrastructural and molecular evidence so far have demonstrated that at least two different lineages of epibiotic bacteria are associated with symbiontids:
- Rod-shaped sulfur-oxidizing epsilon proteobacteria
- Spherical verrucomicrobia containing a tightly wound thread around a dense DNA core that is capable of rapid discharge (SEM and TEMs shown below to the right of the orange cell).
Epsilon proteobacteria have a significant role in deep-sea habitats as primary colonizers, primary producers and/or in symbiotic associations, so these epibionts might detoxify the immediate environment for the eukaryotic hosts. The spherical verrucomicrobia, by contrast, are capable of rapid evisceration of the chromosome and thread through an apical pore. The features of these bacteria are enigmatic but provide context for inferring lateral symbiont transfer, convergent evolution, and potentially the origins of extrusomes in some groups of eukaryotes. The eukaryotic hosts likely serve as a motile substrate that delivers the epibionts to ideal locations with respect to the oxic/anoxic interface, perhaps allowing the host to cultivate a food source.
We are interested in gaining much deeper insights into the overall diversity and basic biology of these particular hosts and epibionts. Because symbiontid isolates and environmental DNA sequences from this clade have now been recovered from many locations worldwide, symbiontids are expected to be much more widespread and diverse than presently appreciated. Projects in the lab will continue to address this diversity using a combination of comparative ultrastructural and molecular approaches, including single cell genomics/transcriptomics.